
MEGSIM MRIView //
MRIVIEW: An Interactive Tool for Brain Imaging
MRIVIEW is a software tool for viewing and manipulating volumetric MRI head data, and for using this data as an anatomical reference in studies of brain function. MRIVIEW supplies methods for reading in raw MRI data, viewing this data in either 2 or 3 dimensions, segmenting structures in the data, reconciling coordinate systems between the MRI data volume and data obtained from brain functional modalities, and viewing combinations of anatomical and functional information. MRIVIEW has three basic operating modes: a two dimensional (2D) mode for viewing and segmenting MRI slice data; a restricted three dimensional (3D) mode used for coordinate reconciliation; and a 3D model viewing mode. For more detail on using MRIVIEW, see the help document: mriview_help.html
2D Viewing Mode
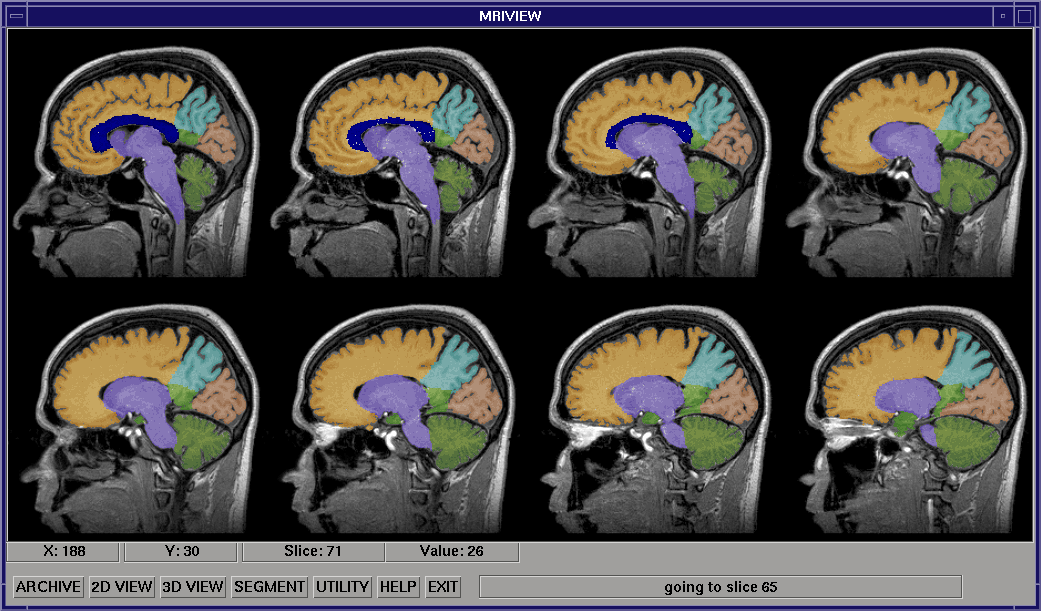
In the 2D mode, the MRI data volume is viewed as a series of slice images in one of the three standard orientations (sagittal, coronal, or axial), which the user can page through, eight at a time. A wide range of structure segmentation methods can be accessed while operating in the two-dimensional mode. These segmentation methods range from user-guided slice by-slice flooding for labelling structures of interest, to an automatic volumetric method incorporating 3D morphological and flooding operations. The image above shows eight slices of an MRI head volume after the brain has been segmented with the automated segmentation procedure, then further segmented using the multi-object segmentation interface.
3D Viewing Mode
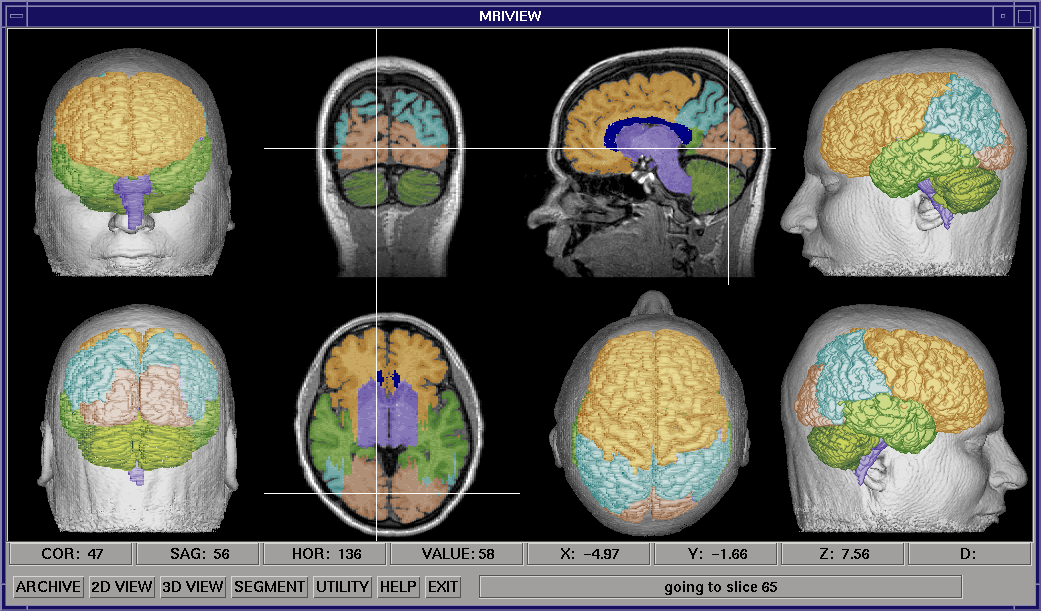
In the restricted 3D mode, a surface obtained by setting an intensity threshold in the MRI data (isosurface) is viewed in five standard orientations, using either a light-shaded rendering (shown at right), or a depth-shaded rendering. These surfaces are displayed with the orthogonal slice views of the MRI data. Selecting a point with a cursor on any of these surfaces will locate and display the corresponding point in the MRI data volume. Using this capability, the fiducial marks used in a functional brain study can be located in the MRI data, and a transformation can be constructed between the two coordinates systems. The image above shows the nasion being selected using the front view of the head. Facilities are provided for using this transformation to plot and print locations of brain function on the MRI data, including an automated method that uses an input source location file. An oblique volume slicer can also be accessed in this viewing mode.
Other Utilities
The Archive submenu provides utilities for reading in MRI files from several machines, as well as a generalized, customizable method for reading files from other MRI or CT machines. Once read into MRIVIEW, the data is in a volume model format, and can be saved to and loaded from files in this form. MRIVIEW also provides utilities for color table manipulation, zooming in on slice images, and annotating the images in the viewing window.
Model Viewer
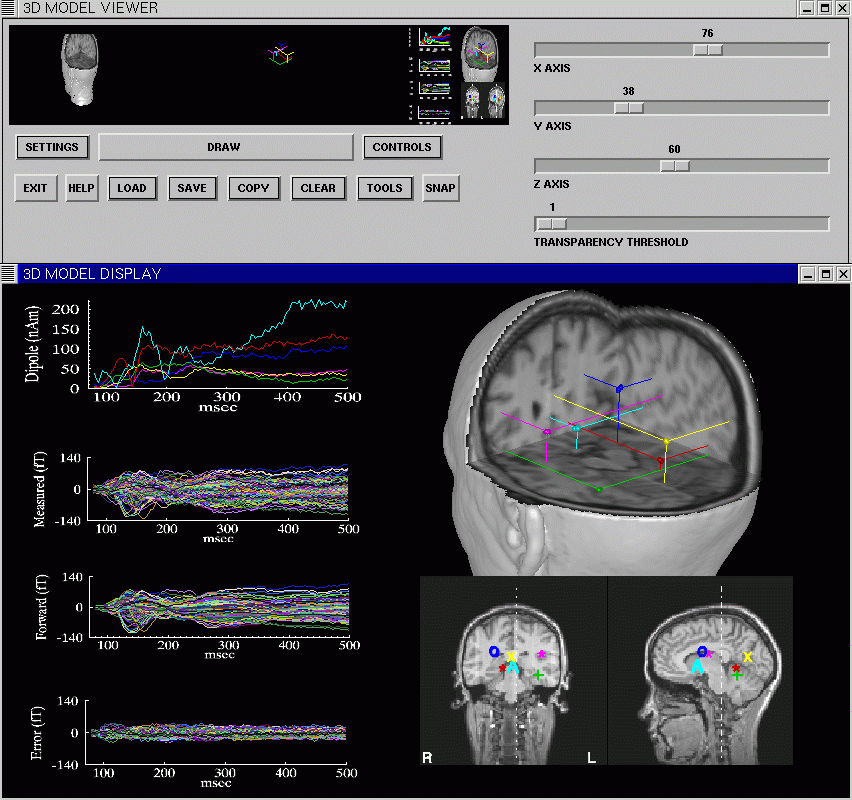
A 3D Model Viewer subsystem of MRIVIEW provides methods for generalized 3D viewing and manipulation of MRI data volumes, and for combining these volumes with geometric models, or volumes containing functional data. The user interface of the Model Viewer consists of a resizable graphics window, and a user interface window. The user can rapidly manipulate low resolution versions of volume models and geometries, then have the program draw a high resolution rendering of the models and geometries. The display properties of each of these objects, such as color and rendering method used, can be manipulated independently. A variety of methods are provided for creating combined displays of MRI volumes and geometric models. The Model Viewer can be used to make movies of objects in the system while they are being systematically rotated, or cut by a moving cutplane. The image above shows the Model Viewer being used in conjunction with the Source Location Plotting interface, to display a multi-dipole solution derived from MEG visual data (using CSST, see below). The MRIVIEW Image Gallery contains several images generated using the 3D Model Viewer.
MEG and EEG Inverse Procedure
A multi-dipole spatial-temporal MEG and EEG inverse precedure is integrated with the 3D Model Viewer, and the Source Location Plotting interface. The Cortical Start Spatial-Temporal (CSST) inverse is a multi-start procedure based on MSST (see Huang et al. Multi-start Downhill simplex Method for Spatio-temporal Source Localization. Magnetoencephalography, Electroencephalography & Clinical Neurophysiology, Vol. 108/1, pp.32-34, Jan. 1998).
In CSST, the MRI data is used to derive the start locations from which the the multiple starts are drawn. For each of these multiple starts, a nonlinear simplex procedure coupled to a singular value decomposition-based timecourse fitting procedure is used to minimize a reduced Chi square value for the user specified model order. CSST can be run as a normal IDL program, and it has also been parallelized for use on Linux clusters. The parallel version (MPI_CSST) requires software not currently distributed with the MRIVIEW installation (available on request). CSST currently relies on MEGAN to produce netMEG files containing MEG or EEG sensor data.
MEG and EEG Forward Simulator
An MEG and EEG forward simulator has been integrated for use with the restricted 3D viewing mode above. The user can specify size, shape, and orientation of ellipsoid-constrained cortical patches, assign a variety of timecourses to these patches, and generate forward models based on a sensor geometry contained in a netMEG file. The simulator outputs netMEG files suitable for use with CSST. Dipole timecourse files, containing mean patch locations and orientations, can also be generated for use with the Source Location Plotting interface.
Other Utilities
The Archive submenu provides utilities for reading in MRI files from several machines, as well as a generalized, customizable method for reading files from other MRI or CT machines. Once read into MRIVIEW, the data is in a volume model format, and can be saved to and loaded from files in this form. MRIVIEW also provides utilities for color table manipulation, zooming in on slice images, and annotating the images in the viewing window.
Computing Platform
MRIVIEW is written in IDL (Interactive Data Language), a product of Research Systems, Inc. IDL is a scientific programming language, and provides a wide range of tools for data analysis and visualization, as well as facilities for developing graphical user interfaces. These tools work on UNIX workstations, and PC and Macintosh platforms. IDL has a data parallel syntax, enabling manipulation of multi-dimensional arrays using arithmetic and logic operators. IDL provides an efficient interactive mode for exploration and prototyping as well as compiled operation for increased efficiency.
Integration and Availability
The use of IDL allows quick integration of MRIVIEW with other software developed here and elsewhere by allowing simple links to other packages written in IDL, C, Fortran, and MATLAB. Currently, MRIVIEW has file based links to Brainstorm and Freesurfer. Future development will include: expanding the inverse analysis capabilities, including incorporating a BEM forward model, and a distributed source analysis; adding at least basic fMRI analysis procedures; and more integration with other software packages. The continuing development of MRIVIEW is supported by the NIH Human Brain Project, and by the MIND Institute.
MRIVIEW is available via anonymous ftp. Source code is not provided with the distribution. Because MRIVIEW is written in IDL, an IDL license is required to use the software. IDL Licensing information can be obtained from Research Systems.

